
Studying environmental DNA and ocean currents to locate fish aggregations in Puerto Rico
- User stories
Synopsis
Challenge
eDNA can provide insights on what types of fish and other organisms are present in a marine ecosystem, but understanding where these trace DNA elements came from is tricky.
Solution
Using an AWAC, researchers from Isla Mar Research Expeditions in Puerto Rico were able to collect current and wave information simultaneously to eDNA samples to better understand eDNA residency time in the water and investigate where it may have originated.
Benefit
The team now has more insights on fish spawning aggregations (a group of fish of the same species that are gathered together for the purpose of spawning) and will be able to monitor them more closely in the future.
Isla Mar Research Expeditions is a field-based marine education and research company founded and led by two fish biologists in Puerto Rico whose primary investigative focus is fisheries management.
Their mission is to provide marine science education, field-based research training and experience and public outreach in ocean conservation throughout Puerto Rico and other parts of the Caribbean.

We had a chat with Fish Biologist Dr. Chelsea Harms-Tuohy about how the group has been using Nortek’s AWAC ADCP as part of their research.
The project, funded by NOAA’s Saltonstall-Kennedy grant program, is a pilot project that investigates the possibility of using eDNA – that is, environmental DNA, which is DNA of organisms that can be extracted from environmental samples – as a means for locating elusive fish spawning aggregations.
These aggregations are generally difficult to locate, as doing so requires the observer to be present at exactly the right time and place. Thus, in an attempt to alleviate some of the traditionally field-intensive search-and-locate methodology, Isla Mar Research Expeditions scientists are piloting a method of collecting water samples at known fish spawning aggregation sites in Puerto Rico to see if they can detect the presence of their target fish.
What was the main focus of your eDNA sampling and fish aggregation research project, where you used a Nortek ADCP?
“This project was our response to the call to locate new fish spawning aggregations in Puerto Rico, with the goal of being able to monitor these aggregations for the future."
“This is no simple task, as locating aggregations requires cooperation with fishers, hours of diving and being in ‘the right place at the right time’, and can be equipment intensive (using passive acoustic devices to monitor for courtship-associated sounds, audible mating sounds that some fish produce that would indicate the potential presence of the target fish). Our research could provide fisheries management (both locally and nationally) with critical information about the timing and duration that a fish spawning aggregation remains in a particular (both spatially and temporally predictable) site. Our goal is to help sustain and replenish our fisheries in Puerto Rico.”

Pardon the ignorance, but what is eDNA sampling?
“eDNA is environmental DNA, or basically DNA that can be detected and obtained from an environmental sample. That sample can be soil, water, air ... DNA of all organisms in that sample can be extracted, or you can just extract DNA from target organisms – which can be a little tricky. In our case, we are searching for a needle in the haystack of targeting one particular fish species in our water samples, but we’re increasing the chances of detecting that target species by sampling during a time that fish is in a greater-than-normal abundance at the sample site.”
Why is your research on eDNA and fish aggregation beneficial for greater society and the advancement of science?
“This is an innovative tool that allows for more rapid assessment and less dive-intensive sampling for our field of fisheries science.
“The ability to detect the presence of fish at a site without actually having to verify that the fish is there is a way for us to rapidly search for unknown spawning aggregation sites. But this is essentially a pilot project, and the verdict is still out on whether this method will work as we hypothesize.
“In the past, we would need to see (or hear, with passive acoustics) the actual fish to know if it was at the spawning site. With eDNA we should be able to detect it was there with or without a visual or auditory record.”

How does this ADCP provide value in this research? What challenges did this instrumentation help you resolve?
“We needed to understand how eDNA could move within and around our ocean environment.
“The ocean being a dynamic system means there are many factors completely outside of our control that can significantly influence our ability to detect the presence of DNA in the water.
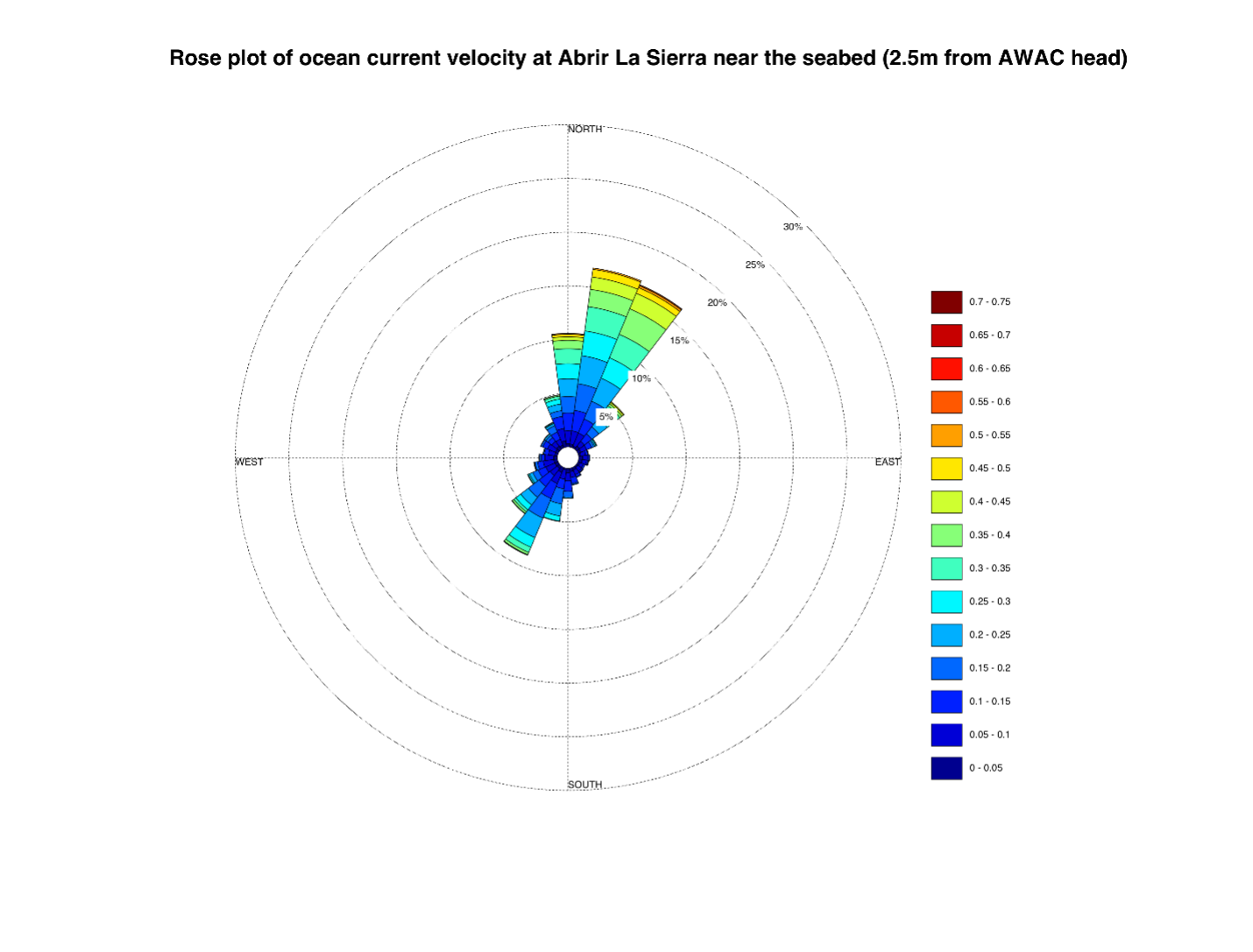
“For example, the speed and direction of the current will determine if we should be upstream or downstream from the fish aggregation at the time that we sample. The AWAC unit was used to tell us what was happening with the current speed and direction at the exact time and place that we collected our water samples at the aggregation site.
“This will help inform us about eDNA residency time in the water and could help us explain reasons for if we detect or cannot detect the presence of DNA in the samples. The ocean is a new environment for eDNA research, and some of these questions are posed by the scientific community, so we hoped to be able to answer those with this project and use of the AWAC ADCP.”
How did the AWAC ADCP perform? Have you got any data that are particularly interesting, useful or outstanding?
“The AWAC helped us to detect when the currents were the strongest and if that occurred while we were sampling. Overall, it was advantageous to be able to see exactly what the ocean was doing at the exact time we were in the water sampling. This will be very helpful in constructing the full story for this project.”

What does the future hold for eDNA research in marine biology and ocean science?
“The future is really open for eDNA application, especially in regard to how it is used in the ocean systems. We plan to use the tool to further explore questions about fish spawning timing and duration, to locate shark mating aggregations and perhaps even to investigate its application for detecting illegal fishing.”
Also check out this ECO Magazine article on how researchers from the National Institute for Environmental Studies, Tohoku University, Shimane University, Kyoto University, Hokkaido University and Kobe University have studied a method for estimating population abundance of fish species by means of measuring concentration of environmental DNA in the water.
In this video from OCEARCH you’ll see researchers using a similar technique to search for the presence of Great White sharks in their eDNA samples.




